|
Recently the European Association for Cancer Research (EACR), who as you know funded me to travel to IARC in Lyon to work, launched a blog competition. The idea of the competition was to write a short blog-style post about "Life in the Lab". You were allowed to submit as many blogs as you liked so I submitted three:
I found out today that the first one (Engaging with the (scary) public – why we need to do it and do it now) was shortlisted as one of the best blogs submitted. So while I didn't win, it will still get published in the next few months. BUT what I also learnt was that they liked my other two so much that they are going to published anyway probably early next year! How awesome is that! Since they are going to be published I won't be adding them up here but watch this space. Once they become available on the EACR magazine website I will let you all know!
0 Comments
Yesterday I was at the New Scientist Live exhibition in the ExCEL London with CRUK (sorry I am terrible at taking photos so eh no evidence). My job there was to chat to the future scientists from 13 year olds to 23 year olds about how to get a career in research and what it was like to do a PhD. And my one biggest piece of advice was do coding! Bioinformatics is the future. Seriously, even if you hate maths (like me) learning some code language will help you in anything you do.
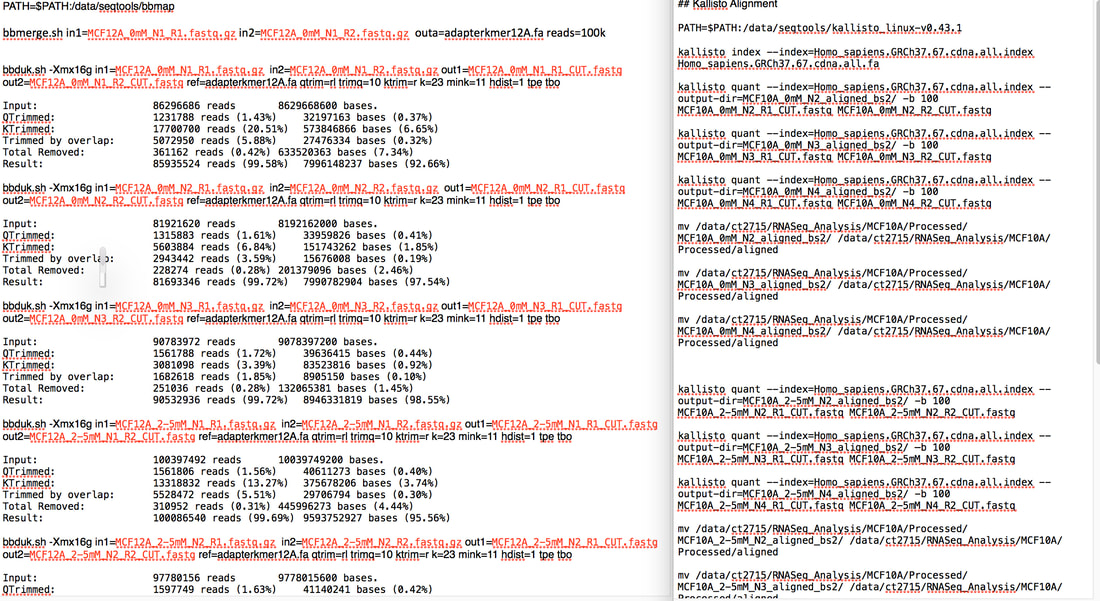
My other piece of advice was that if you want a PhD google www.findaphd.com/ because it does exactly what it says on the tin. It found me a PhD. So what is bioinformatics and why is it important? Bioinformatics is basically the analysis of biological data. Biological data usually means sequencing data. You use a coding language such as R, python or unix to analyse this data. The above picture is my code for doing quality control on my RNA sequencing data using unix. I can also (barely) use R. To me python is a snake... Bioinformatics is important because we can take massive amounts of data from hundreds or thousands of different samples and use this to generate more knowledge about areas such as cancer. This can be sequencing the entire compliment of genes in tumours of hundreds of patients to see if there are common mutations. This can tell us what these tumours have in common and what they don't. You can use this information for example to find "driver mutations" which are mutations commonly found in early stage tumours (p53 mutation is a driver mutation as some cancers require loss of p53 to develop into cancer). The idea of using whole genome sequencing in cancer has come up a lot recently in the news. And the people who are taking this data and finding interesting results are bioinformaticians. Pretty much every cancer research lab in the world is generating data which needs to be analysed computationally. The data you get collectively from sequencing etc. is called "Big Data". However while most labs are generating this data, a large amount of those labs have no one to analyse it. We need more bioinformaticians to take this data and make it useful. Hence why bioinformatics is the future. The dawn of the era of joint experimental and computational scientists is dawning. I am an experimental scientist (i.e I could barely use excel let alone do big data analysis when I started my PhD). But I had to learn how to use R and unix to analyse my data because frankly there was no one else. And this learning was something actively encouraged by my supervisor. Every PhD and post doc James has now can do both. It's very important to him. It really should be more important to a lot more supervisors. So here's my experience: I have done two types of sequencing experiments generating data on DNA methylation (Illumina MethylationEPIC Array) AND gene expression data (RNA-Seq) of two non-cancerous cells treated with metformin. What this gives me in a massive amount of raw data (like hundreds and hundreds of gigabytes - the total amount of data generated from my RNA-Seq was roughly 2-3 terabytes...yeah). I took this raw data and ran it through quality control to make sure every piece of information I got is the best quality. I then ran analyses to determine if there are differences between DNA methylation levels or gene expression levels between your untreated and treated samples. For example if I see a consistent, large reduction in DNA methylation in a certain gene that suggests that there is more gene expression at that gene (low methylation = expression, high methylation = no expression). I can actually overlay this with my RNA sequencing data to see if that gene does actually increase expression! It's pretty cool. I mean you have to validate everything you see in your sequencing data with specific experiments but you can find some very interesting things. So what was the catch (besides learning how to make my computer do this stuff)? It takes time. It took me a full year to analyse my DNA methylation data and validate what I found. I have only started analysing my RNA-Seq data and I can tell you I have sat for 7 hours today watching my computer do aligning on my samples (it's only done 5 out of 9). And god forbid I put a comma in the wrong place or didn't add some arbitrary bit of syntax! I honestly spent a full day trying to figure out why the code I had written didn't work after the computer kept spitting out the "how to" file for the function I had used. I had done exactly what the were saying...except I didn't add two dashes, I had only added one. A bloody dash. My take home message is the more information we have the more we know. Seems simple right? The gathering of that data is in the whole pretty easy to be honest and a lot cheaper than it once was (though both of my sequencing experiments cost a total of around £9,000 - £10,000 each to get the data). The challenge is analysing that data. Handling terabytes of data to find even one gene shows a change in expression is a mammoth task. We need more people who can pick up a pipette and run an experiment as well as analyse the results of that experiment. If you are interested in learning any coding language there are lots of resources online as well as courses you can do. My supervisor (James Flanagan) and a fellow bioinformatician (he's a genius - Ed Curry) run a masters in Imperial in cancer informatics. It may seem daunting but if I can learn it so can you. And not just for cancer research or science in general. Knowing how to code could benefit a lot of industries. Bioinformatics! It's the future people! Sure how are you going to programme your robot in the future if you don't know code?? Resources I found useful: www.youtube.com/user/marinstatlectures www.tutorialspoint.com/r/ www.coursera.org/learn/data-scientists-tools www.imperial.ac.uk/study/pg/medicine/cancer-informatics/ |
AuthorMy name is Caitriona and I am a PhD student at Imperial College London, UK. Categories
All
|